Persistent Volumes
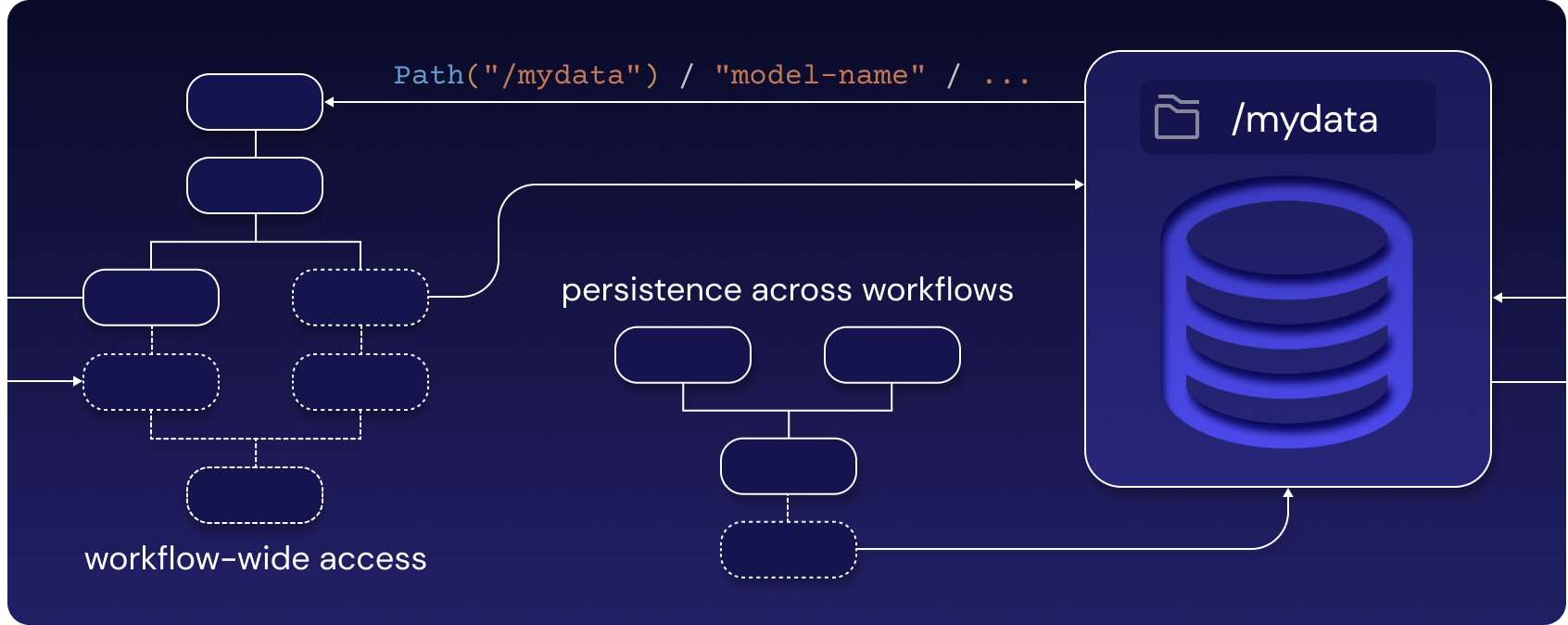
While each task has access to temporary storage at runtime, no persistent storage is available to workflow tasks by default. Covalent Cloud provides a simple interface enabling persistent storage with cc.volume(). This function creates a new volume (or refers to an existing volume) which can be attached to any workflow and/or function service during dispatch.
my_volume = cc.volume("my-volume")
runid = cc.dispatch(my_workflow, volume=volume)(*args, **kwargs)
This lets any task inside workflow read or write to files in /volumes/my_volume.
Why use volumes?
Relying on task inputs and outputs is not generally recommended for transferring very large amounts of data. Task inputs and outputs are in fact limited to 2.5 Gb in Covalent Cloud. A much better alternative here is to attach a storage volume to your workflows.
Volumes in Covalent Cloud belong to a user, rather than a particular workflow or function service. This means that volumes store data indefinitely, until explicit deletion. Any volume can be attached to any workflow or function service.
When attached to workflows, a volume is accessible workflow-wide (as shared storage across tasks). When attached to function services, a volume becomes accessible to all endpoints, as well as the service initializer.
Volumes are also useful for avoiding repeated downloads across concurrent workflow tasks.
Using volumes with workflows
Let’s take a look at a simple example.
import covalent as ct
import covalent_cloud as cc
from datetime import datetime
cc.save_api_key("your-api-key")
my_volume = cc.volume("/my_volume") # creates the volume if it doesn't exist
Consider the following task, which creates specific files when it executes. The volume my_volume is used to store these files.
ex = cc.CloudExecutor(env="my-env")
@ct.electron(executor=ex)
def create_new_files(label):
# Create directory if it does not exist.
label_dir = my_volume / label
label_dir.mkdir(exist_ok=True)
# Write to new log file on volume.
timestamp = datetime.now().strftime("%H-%M-%S")
new_log_file = label_dir / f"log_{timestamp}.txt"
new_log_file.write_text(f"This file was created at {timestamp}.\n")
return label_dir
This next task reads data from files on the volume.
@ct.electron(executor=ex)
def read_existing(log_dirs):
log_contents = {}
# Read log files in the directory.
for log_dir in log_dirs:
for log_file in log_dir.glob("log_*.txt"):
log_contents[str(log_file)] = log_file.read_text()
return log_contents
Now, let’s create a simple workflow to do read and write a number of files.
@ct.lattice(executor=ex, workflow_executor=ex)
def workflow(*labels):
"""Manipulates model files, performs updates, and reads logs."""
log_dirs = []
for label in labels:
log_dirs.append(create_new_files(label)) # parallel tasks
return read_existing(log_dirs)
Dispatching with volumes attached
With the inputs below, subsequent dispatches will reveal three more files each time. Let’s quickly confirm with two sequential dispatches.
from pprint import pprint
dispatch_id = cc.dispatch(workflow, volume=my_volume)( # volume attached here
"model-1a-small",
"model-3c-small",
"model-3c-large"
)
result = cc.get_result(dispatch_id, wait=True).result
result.load()
for k ,v in result.value.items():
print(f"File: {k}\nContents: {v}")
Here’s what we get the first time we dispatch this workflow.
File: /volumes/my-volume/model-1a-small/log_19-06-48.txt
Contents: This file was created at 19-06-48.
File: /volumes/my-volume/model-3c-small/log_19-06-48.txt
Contents: This file was created at 19-06-48.
File: /volumes/my-volume/model-3c-large/log_19-07-09.txt
Contents: This file was created at 19-07-09.
Dispatching a second time indeed reveals three additional files:
File: /volumes/my-volume/model-1a-small/log_19-06-48.txt
Contents: This file was created at 19-06-48.
File: /volumes/my-volume/model-1a-small/log_19-09-15.txt
Contents: This file was created at 19-09-15.
File: /volumes/my-volume/model-3c-small/log_19-06-48.txt
Contents: This file was created at 19-06-48.
File: /volumes/my-volume/model-3c-small/log_19-08-59.txt
Contents: This file was created at 19-08-59.
File: /volumes/my-volume/model-3c-large/log_19-07-09.txt
Contents: This file was created at 19-07-09.
File: /volumes/my-volume/model-3c-large/log_19-08-59.txt
Contents: This file was created at 19-08-59.
Using volumes with function services
Like workflows, function services can also access persistent storage in Covalent Cloud. Passing a volume to the @cc.service decorator makes the volume accessible to both the initializer and all endpoint functions comprising the service.
@cc.service(executor=service_executor, volume=my_volume) # attach volume here
def example_service():
log_file = my_volume / "log_file.txt"
with open(log_file, "w") as log:
log.write("This file was created by a service initializer.\n")
return {"init_log": log_file}
When using in-workflow deployments, a service’s volume (specified in @cc.service) is fully independent from the volume of its parent workflow (specified in @cc.dispatch). Each must be specified individually.